- PrognoScan: A new database for meta-analysis of the prognostic value of genes.
manuscript - presentation
PrognoScan searches the relation between gene expression and patient progonsis such as overall survival (OS) and disease free survival (DFS) across a large collection of publicly available cancer microarray datasets.
- Background
In cancer research, the association between a gene and clinical outcome suggests the underlying etiology of the disease and consequently can motivate further studies. The recent availability of published cancer microarray datasets with clinical annotation provides the opportunity for linking gene expression to prognosis. However, the data are not easy to access and analyze without an effective analysis platform.
- Description
To take advantage of public resources in full, a database named "PrognoScan" has been developed.
This is 1) a large collection of publicly available cancer microarray datasets with clinical annotation,
as well as 2) a tool for assessing the biological relationship between gene expression and prognosis.
PrognoScan employs the minimum P-value approach for grouping patients for survival analysis that finds the optimal cutpoint in continuous gene expression measurement without prior biological knowledge or assumption and, as a result, enables systematic meta-analysis of multiple datasets.
- Conclusion
PrognoScan provides a powerful platform for evaluating potential tumor markers and therapeutic targets and would accelerate cancer research. The database is publicly accessible at http://www.prognoscan.org/.
- Screenshots
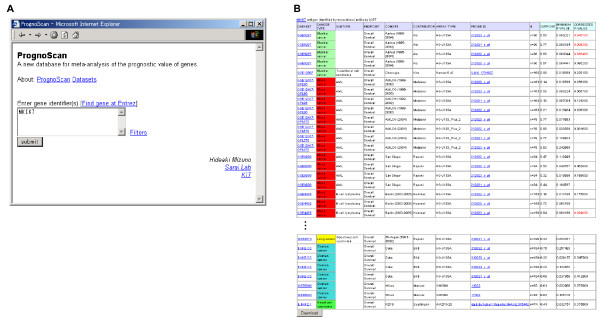
PrognoScan screenshot and sample search results (part 1).
(A) The top page is quite simple and only requires entering the gene identifier(s). (B) Summary table. Column headings include dataset, cancer type, subtype, endpoint, cohort, contributor, array type, probe ID, number of patients, optimal cutpoint, Pmin and Pcor. A statistically significant value of Pcor is given in red font. Each dataset has a link to the public domain where the raw data is archived. By clicking a probe ID in the summary table, a detailed report for the test is displayed. The table can be downloaded in a tab delimited file from the button at bottom.
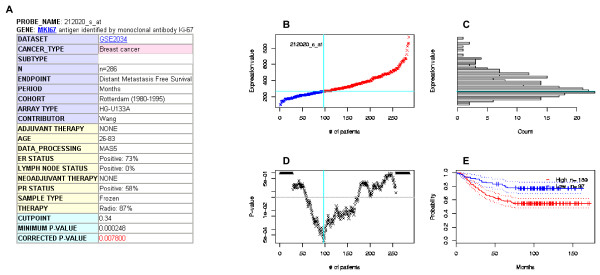
PrognoScan screenshot and sample search results (part 2).
(A) Annotation table. Row headings are color-coded. For example, headings of details such as therapy history, sample type and pathological parameters are highlighted in yellow and basic attributes in blue. (B) Expression plot. Patients are ordered by the expression values of the given gene. The X-axis represents the accumulative number of patients and the Y-axis represents the expression value. Straight lines (cyan) show the optimal cutpoints that dichotomize patients into high (red) and low (blue) expression groups. (C) Expression histogram. The distribution of the expression value is presented where the X-axis represents the number of patients and the Y-axis represents the expression value on the same scale as the expression plot. The line of the optimal cutpoint is also shown (cyan). (D) P-value plot. For each potential cutpoint of expression measurement, patients are dichotomized and survival difference between high and low expression groups is calculated by log-rank test. The X-axis represents the accumulative number of patients on the same scale as the expression plot and the Y-axis represents raw P-values on a log scale. The cutpoint to minimize the P-value is determined and indicated by the cyan line. The gray line indicates the 5% significance level. (E) Kaplan-Meier plot. Survival curves for high (red) and low (blue) expression groups dichotomized at the optimal cutpoint are plotted. The X-axis represents time and the Y-axis represents survival rate. 95% confidence intervals for each group are also indicated by dotted lines.
- Newsletter
2012/09/17 The stable address http://www.prognoscan.org has been set.
2011/11/16 The Okayama et al. dataset of lung adenocarcinomas has been added.
2011/11/16 The Gobble et al. dataset of liposarcoma has been added.
2011/08/27 The Laurent et al. dataset of uveal melanomas has been added.
2011/08/22 The Bonome et al. dataset of ovarian cancers has been added.
2011/06/22 The Wilkerson et al. dataset of lung squamous cell carcinomas has been added.
2011/04/13 The Zhu et al. dataset of non-small cell lung cancers has been added.
2011/04/13 The Lee et al. dataset of meningiomas has been added.
2011/01/29 The Bos et al. dataset of breast cancers has been added.
2011/01/10 The Denkert et al. dataset of ovarian cancers has been added.
2010/06/28 P-values and HRs from Cox univariate analysis have been added.
2010/06/28 PrognoScan has adopted original processed expression values as default setting.
2010/06/28 The Yoshihara et al. dataset of ovarian cancers has been added.
2010/06/28 The Smith et al. dataset of colorectal cancers has been added.
2010/06/28 The Li et al. dataset of breast cancers has been added.
2010/06/28 The Nutt et al. dataset of gliomas has been added.
2010/06/28 The Freije et al. dataset of gliomas has been added.
2010/06/28 The Sboner et al. dataset of prostate cancers has been added.
2010/02/10 The Jorissen et al. dataset of colorectal cancers has been added.
2009/12/23 The Shedden et al. dataset of lung adenocarcinomas has been added.
2009/12/23 The Bogunovic et al. dataset of metastatic melanomas has been added.
2009/12/05 An option for reporting test results based on original processed data has been added.
2009/11/13 A column for hazard ratio (HR) between high and low expression groups has been added.
2009/10/10 The Dave et al. dataset of follicular lymphomas has been added.
2009/08/09 The Tomida et al. dataset of lung adenocarcinomas has been added.
2009/06/13 The Staub et al. dataset of colorectal cancers has been added.
2009/04/24 PrognoScan has been published in BMC Med Genomics. 2009 2:18.
- Bibliography
The PrognoScan database has been referred in
- H3K9me3 facilitates hypoxia-induced p53-dependent apoptosis through repression of APAK. Olcina MM, Leszczynska KB, Senra JM, Isa NF, Harada H, Hammond EM. Oncogene. 2015.
- Integrative genomic analyses of a novel cytokine, interleukin-34 and its potential role in cancer prediction. Wang B, Xu W, Tan M, Xiao Y, Yang H, Xia TS. Int J Mol Med. 2015 35:92-102.
- CRKL oncogene is downregulated by p53 through miR-200s. Tamura M, Sasaki Y, Kobashi K, Takeda K, Nakagaki T, Idogawa M, Tokino T. Cancer Sci. 2015.
- A favourable prognostic marker for EGFR mutant non-small cell lung cancer: immunohistochemical analysis of MUC5B. Wakata K, Tsuchiya T, Tomoshige K, Takagi K, Yamasaki N, Matsumoto K, Miyazaki T, Nanashima A, Whitsett JA, Maeda Y, Nagayasu T. BMJ Open. 2015 5:e008366.
- An Arf-Egr-C/EBPβ pathway linked to ras-induced senescence and cancer. Salotti J, Sakchaisri K, Tourtellotte WG, Johnson PF. Mol Cell Biol. 2015 35:866-83.
- Inhibitory Mechanism of FAT4 Gene Expression in Response to Actin Dynamics during Src-Induced Carcinogenesis. Ito T, Taniguchi H, Fukagai K, Okamuro S, Kobayashi A. PLoS One. 2015 10:e0118336.
- The AURKA/TPX2 axis drives colon tumorigenesis cooperatively with MYC. Takahashi Y, Sheridan P, Niida A, Sawada G, Uchi R, Mizuno H, Kurashige J, Sugimachi K, Sasaki S, Shimada Y, Hase K, Kusunoki M, Kudo S, Watanabe M, Yamada K, Sugihara K, Yamamoto H, Suzuki A, Doki Y, Miyano S, Mori M, Mimori K. Ann Oncol. 2015 26:935-42.
- UCHL1 provides diagnostic and antimetastatic strategies due to its deubiquitinating effect on HIF-1α. Goto Y, Zeng L, Yeom CJ, Zhu Y, Morinibu A, Shinomiya K, Kobayashi M, Hirota K, Itasaka S, Yoshimura M, Tanimoto K, Torii M, Sowa T, Menju T, Sonobe M, Kakeya H, Toi M, Date H, Hammond EM, Hiraoka M, Harada H. Nat Commun. 2015 6:6153.
- Survival analysis tools in genomics research. Chen X, Sun X, Hoshida Y. Hum Genomics. 2014 8:21.
- PROGgeneV2: enhancements on the existing database. Goswami CP1, Nakshatri H. BMC Cancer. 2014 14:970.
- Integrative genomic analyses of a novel cytokine, interleukin-34 and its potential role in cancer prediction. Wang B, Xu W, Tan M, Xiao Y, Yang H, Xia TS. Int J Mol Med. 2015 35:92-102.
- Aberrant IDH3α expression promotes malignant tumor growth by inducing HIF-1-mediated metabolic reprogramming and angiogenesis. Zeng L, Morinibu A, Kobayashi M, Zhu Y, Wang X, Goto Y, Yeom CJ, Zhao T, Hirota K, Shinomiya K, Itasaka S, Yoshimura M, Guo G, Hammond EM, Hiraoka M, Harada H. Oncogene. 2014
- Survival analysis tools in genomics research. Chen X, Sun X, Hoshida Y. Hum Genomics. 2014 8:21.
- ISG15 Is a Critical Microenvironmental Factor for Pancreatic Cancer Stem Cells. Sainz B Jr, Martín B, Tatari M, Heeschen C, Guerra S. Cancer Res. 2014 74:7309-20.
- Downregulation of PRRX1 Confers Cancer Stem Cell-Like Properties and Predicts Poor Prognosis in Hepatocellular Carcinoma. Hirata H, Sugimachi K, Takahashi Y, Ueda M, Sakimura S, Uchi R, Kurashige J, Takano Y, Nanbara S, Komatsu H, Saito T, Shinden Y, Iguchi T, Eguchi H, Atsumi K, Sakamoto K, Doi T, Hirakawa M, Honda H, Mimori K. Ann Surg Oncol. 2014.
- Epithelial Splicing Regulatory Proteins 1 (ESRP1) and 2 (ESRP2) Suppress Cancer Cell Motility via Different Mechanisms. Ishii H, Saitoh M, Sakamoto K, Kondo T, Katoh R, Tanaka S, Motizuki M, Masuyama K, Miyazawa K. J Biol Chem. 2014 289:27386-99.
- Integrative genomic analyses of secreted protein acidic and rich in cysteine and its role in cancer prediction. Wang B, Chen K, Xu W, Chen D, Tang W, Xia TS. Mol Med Rep. 2014 10:1461-8.
- Analysis of acquired resistance to metronomic oral topotecan chemotherapy plus pazopanib after prolonged preclinical potent responsiveness in advanced ovarian cancer. Cruz-Muñoz W, Di Desidero T, Man S, Xu P, Jaramillo ML, Hashimoto K, Collins C, Banville M, O'Connor-McCourt MD, Kerbel RS. Angiogenesis. 2014 17:661-73.
- Integrative genomic analyses of the histamine H1 receptor and its role in cancer prediction. Wang M, Wei X, Shi L, Chen B, Zhao G, Yang H. Int J Mol Med. 2014 33:1019-26.
- Constitutive IDO expression in human cancer is sustained by an autocrine signaling loop involving IL-6, STAT3 and the AHR. Litzenburger UM, Opitz CA, Sahm F, Rauschenbach KJ, Trump S, Winter M, Ott M, Ochs K, Lutz C, Liu X, Anastasov N, Lehmann I, Höfer T, von Deimling A, Wick W, Platten M. Oncotarget. 2014 5:1038-51.
- TSPAN2 Is Involved in Cell Invasion and Motility during Lung Cancer Progression. Otsubo C, Otomo R, Miyazaki M, Matsushima-Hibiya Y, Kohno T, Iwakawa R, Takeshita F, Okayama H, Ichikawa H, Saya H, Kiyono T, Ochiya T, Tashiro F, Nakagama H, Yokota J, Enari M. Cell Rep. 2014 7:527-38.
- Widespread FRA1-Dependent Control of Mesenchymal Transdifferentiation Programs in Colorectal Cancer Cells. Diesch J, Sanij E, Gilan O, Love C, Tran H, Fleming NI, Ellul J, Amalia M, Haviv I, Pearson RB, Tulchinsky E, Mariadason JM, Sieber OM, Hannan RD, Dhillon AS. PLoS One. 2014 9:e88950.
- Cell division cycle-associated protein 1 overexpression is essential for the malignant potential of colorectal cancers. Kobayashi Y1, Takano A, Miyagi Y, Tsuchiya E, Sonoda H, Shimizu T, Okabe H, Tani T, Fujiyama Y, Daigo Y. Int J Oncol. 2014 44:69-77.
- Defining the protein-protein interaction network of the human hippo pathway. Wang W, Li X, Huang J, Feng L, Dolinta KG, Chen J. Mol Cell Proteomics. 2014 13:119-31.
- Metabolic and functional genomic studies identify deoxythymidylate kinase as a target in LKB1-mutant lung cancer. Liu Y, Marks K, Cowley GS, Carretero J, Liu Q, Nieland TJ, Xu C, Cohoon TJ, Gao P, Zhang Y, Chen Z, Altabef AB, Tchaicha JH, Wang X, Choe S, Driggers EM, Zhang J, Bailey ST, Sharpless NE, Hayes DN, Patel NM, Janne PA, Bardeesy N, Engelman JA, Manning BD, Shaw RJ, Asara JM, Scully R, Kimmelman A, Byers LA, Gibbons DL, Wistuba II, Heymach JV, Kwiatkowski DJ, Kim WY, Kung AL, Gray NS, Root DE, Cantley LC, Wong KK. Cancer Discov. 2013 3:870-9.
- RanGTPase: a candidate for Myc-mediated cancer progression. Yuen HF, Gunasekharan VK, Chan KK, Zhang SD, Platt-Higgins A, Gately K, O'Byrne K, Fennell DA, Johnston PG, Rudland PS, El-Tanani M. J Natl Cancer Inst. 2013 105:475-88.
- AKR1B10, a Transcriptional Target of p53, Is Downregulated in Colorectal Cancers Associated with Poor Prognosis. Ohashi T, Idogawa M, Sasaki Y, Suzuki H, Tokino T. Mol Cancer Res. 2013 11:1554-63.
- Germline variation in NCF4, an innate immunity gene, is associated with an increased risk of colorectal cancer. Ryan BM, Zanetti KA, Robles AI, Schetter AJ, Goodman J, Hayes RB, Huang WY, Gunter MJ, Yeager M, Burdette L, Berndt SI, Harris CC. Int J Cancer. 2013 134:1399-407.
- Forkhead transcription factor FOXF1 is a novel target gene of the p53 family and regulates cancer cell migration and invasiveness. Tamura M, Sasaki Y, Koyama R, Takeda K, Idogawa M, Tokino T. Oncogene. 2013.
- PROGgene: gene expression based survival analysis web application for multiple cancers. Goswami CP, Nakshatri H. J Clin Bioinforma. 2013 3:22.
- SurvExpress: an online biomarker validation tool and database for cancer gene expression data using survival analysis. Aguirre-Gamboa R, Gomez-Rueda H, Martínez-Ledesma E, Martínez-Torteya A, Chacolla-Huaringa R, Rodriguez-Barrientos A, Tamez-Peña JG, Treviño V. PLoS One. 2013 8:e74250.
- Targeting the XIAP/caspase-7 complex selectively kills caspase-3-deficient malignancies. Lin YF, Lai TC, Chang CK, Chen CL, Huang MS, Yang CJ, Liu HG, Dong JJ, Chou YA, Teng KH, Chen SH, Tian WT, Jan YH, Hsiao M, Liang PH. J Clin Invest. 2013 123:3861-75.
- Carboxyl-terminal modulator protein positively regulates Akt phosphorylation and acts as an oncogenic driver in breast cancer. Liu YP, Liao WC, Ger LP, Chen JC, Hsu TI, Lee YC, Chang HT, Chen YC, Jan YH, Lee KH, Zeng YH, Hsiao M, Lu PJ. Cancer Res. 2013 73:6194-205.
- C/EBPγ suppresses senescence and inflammatory gene expression by heterodimerizing with C/EBPβ. Huggins CJ, Malik R, Lee S, Salotti J, Thomas S, Martin N, Qui単ones OA, Alvord WG, Olanich ME, Keller JR, Johnson PF. Mol Cell Biol. 2013 33:3242-58.
- Paired related homoeobox 1, a new EMT inducer, is involved in metastasis and poor prognosis in colorectal cancer. Takahashi Y, Sawada G, Kurashige J, Uchi R, Matsumura T, Ueo H, Takano Y, Akiyoshi S, Eguchi H, Sudo T, Sugimachi K, Doki Y, Mori M, Mimori K. Br J Cancer. 2013 109:307-11.
- RASEF is a novel diagnostic biomarker and a therapeutic target for lung cancer. Oshita H, Nishino R, Takano A, Fujitomo T, Aragaki M, Kato T, Akiyama H, Tsuchiya E, Kohno N, Nakamura Y, Daigo Y. Mol Cancer Res. 2013 11:937-51.
- Tumor-derived Tenascin-C Promotes the Epithelial-Mesenchymal Transition in Colorectal Cancer Cells. Takahashi Y, Sawada G, Kurashige J, Matsumura T, Uchi R, Ueo H, Ishibashi M, Takano Y, Akiyoshi S, Iwaya T, Eguchi H, Sudo T, Sugimachi K, Yamamoto H, Doki Y, Mori M, Mimori K. Anticancer Res. 2013 33:1927-34.
- Integrative genomic analyses of recepteur d'origine nantais and its prognostic value in cancer. Yu H, Yuan J, Xiao C, Qin Y. Int J Mol Med. 2013 31:1248-54.
- canEvolve: A Web Portal for Integrative Oncogenomics. Samur MK, Yan Z, Wang X, Cao Q, Munshi NC, Li C, Shah PK. PLoS One. 2013 8:e56228.
- Adenylate Kinase-4 Is a Marker of Poor Clinical Outcomes That Promotes Metastasis of Lung Cancer by Downregulating the Transcription Factor ATF3. Jan YH, Tsai HY, Yang CJ, Huang MS, Yang YF, Lai TC, Lee CH, Jeng YM, Huang CY, Su JL, Chuang YJ, Hsiao M. Cancer Res. 2012 72:5119-29.
- Identification of galanin and its receptor GalR1 as novel determinants of resistance to chemotherapy and potential biomarkers in colorectal cancer. Stevenson L, Allen WL, Turkington R, Jithesh PV, Proutski I, Stewart GE, Lenz HJ, Van Schaeybroeck S, Longley DB, Johnston PG. Clin Cancer Res. 2012 18:5412-26.
- Combinatorial use of bone morphogenetic protein 6, noggin and SOST significantly predicts cancer progression. Yuen HF, McCrudden CM, Grills C, Zhang SD, Huang YH, Chan KK, Chan YP, Wong ML, Law S, Srivastava G, Fennell DA, Dickson G, El-Tanani M, Chan KW. Cancer Sci. 2012 103:1145-54.
- Comprehensive literature review and statistical considerations for microarray meta-analysis. Tseng GC, Ghosh D, Feingold E. Nucleic Acids Res. 2012 40:3785-99.
- Estrogen and antiestrogens alter breast cancer invasiveness by modulating the transforming growth factor-beta signaling pathway. Goto N, Hiyoshi H, Ito I, Tsuchiya M, Nakajima Y, Yanagisawa J. Cancer Sci. 2011 102:1501-8.
- Prediction of survival in diffuse large B-cell lymphoma based on the expression of 2 genes reflecting tumor and microenvironment. Alizadeh AA, Gentles AJ, Alencar AJ, Liu CL, Kohrt HE, Houot R, Goldstein MJ, Zhao S, Natkunam Y, Advani RH, Gascoyne RD, Briones J, Tibshirani RJ, Myklebust JH, Plevritis SK, Lossos IS, Levy R. Blood. 2011 118:1350-8.
- Identification of Genes Up-regulated in ALK-positive and EGFR/KRAS/ALK-negative Lung Adenocarcinomas. Okayama H, Kohno T, Ishii Y, Shimada Y, Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S, Watanabe SI, Sakamoto H, Kumamoto K, Takenoshita S, Gotoh N, Mizuno H, Sarai A, Kawano S, Yamaguchi R, Miyano S, Yokota J. Cancer Res. 2011 72:100-11.
- NONO and RALY proteins are required for YB-1 oxaliplatin induced resistance in colon adenocarcinoma cell lines. Tsofack SP, Garand C, Sereduk C, Chow D, Aziz M, Guay D, Yin HH, Lebel M. Mol Cancer. 2011 10:145.
- An integrative approach to identify YB-1-interacting proteins required for cisplatin resistance in MCF7 and MDA-MB-231 breast cancer cells. Garand C, Guay D, Sereduk C, Chow D, Tsofack SP, Langlois M, Perreault E, Yin HH, Lebel M. Cancer Sci. 2011 102:1410-7.
- Integrative genomic analyses on IL28RA, the common receptor of interferon-lambda1, -lambda2 and -lambda3. Yang L, Luo Y, Wei J, He S. Int J Mol Med. 2010 25:807-12.
- Integrative genomic analyses on Ikaros and its expression related to solid cancer prognosis. Yang L, Luo Y, Wei J. Oncol Rep. 2010 24:571-7.
- Integrative genomic analyses on interferon-lambdas and their roles in cancer prediction. Yang L, Wei J, He S. Int J Mol Med. 2010 25:299-304.
- Notes
Please cite the use of this database as:
PrognoScan: A new database for meta-analysis of the prognostic value of genes. Mizuno H, Kitada K, Nakai K, Sarai A. BMC Med Genomics. 2009 2:18.
Dynamic links to the PrognoScan search results can be created using Entrez gene IDs as:
http://www.abren.net/PrognoScan-cgi/PrognoScan.cgi?MODE=CAL_PROGNOSIS_GID&QUERY=Entrez gene ID
Feedbacks and bug reports are highly appreciated. Please use the following address for contact:
Hideaki Mizuno
- Links
AE: ArrayExpress
GEO: Gene Expression Omnibus
caArray: Array Data Management System
ITTACA: Integrated Tumor Transcriptome Array and Clinical data Analysis
SurvExpress: Biomarker validation for cancer gene expression
PROGgene: Pan Cancer Prognostics Database
LSDB: Life Science DataBase
PrognoScan